Sample Locus Specific Amplification Efficiency (LSAE) according to prior
sample_LSAE.RdSample Locus Specific Amplification Efficiency (LSAE) according to prior
Arguments
- LSAE_variance
Numeric. See gf_configuration for an example.
- locus_names
Character vector.
Details
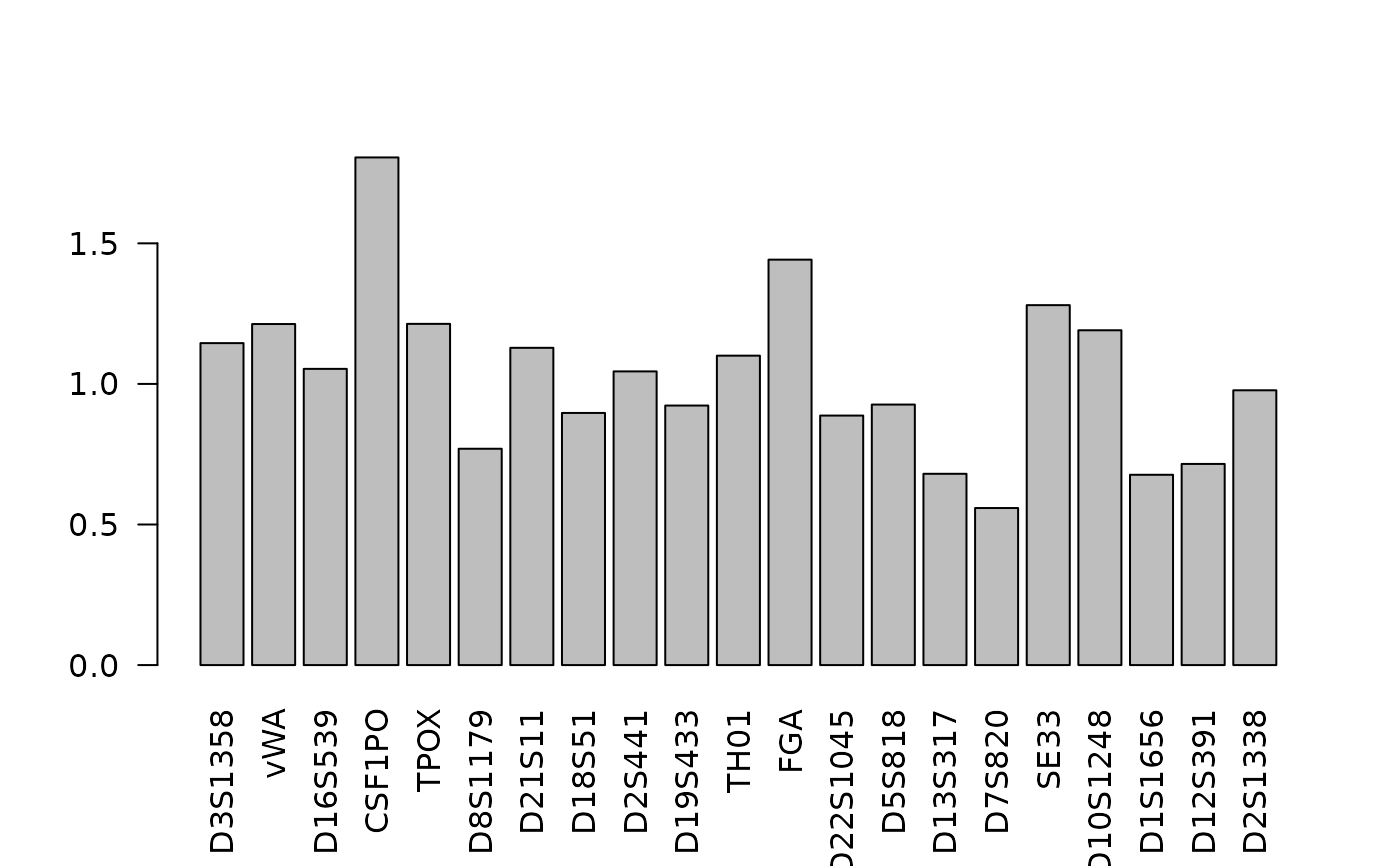
In the Bright et al. log normal model, the expected peak height includes a multiplicative factor for the locus (marker). These factors are called the LSAEs (Locus Specific Amplification Efficiencies). In the model, the prior for the log10 of LSAE is normal with mean 0. The variance can be specified.
Examples
gf <- gf_configuration()
lsae <- sample_LSAE(LSAE_variance = gf$log_normal_settings$LSAE_variance_prior,
locus_names = gf$autosomal_markers)
# the barplot shows that some loci amplify better than others
barplot(lsae, las=2)